
Cluster visualisation is performed similar to CATHAI i.e. GraphSNP generates pairwise Hamming distance from the SNP alignment, visualises it on the network and applies a threshold. These pages are all linked together and managed by the application state management module. The Documentation page (Additional File 1) provides users with a quick start guide and examples of input files.
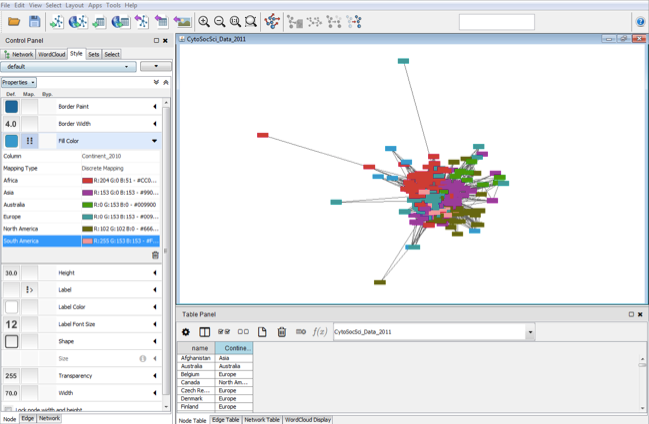
The Distances and Graph pages utilise a graph processing module that allows users to create interactive charts, network, trees and perform cluster and transmission analysis, based on the SNP distance and other related epidemiological data. The Input page is a landing page containing a parser module and input placeholder where users upload their data files. GraphSNP is comprised of four main modules that handle several interconnected pages: Input, Distances, Graph and Documentation (Fig.
#React cytoscape Offline#
Users can access GraphSNP online at or use it offline through a locally-installed web server. The compiled SPA is served using a web server and is accessed via an internet browser. GraphSNP implements graph models and algorithms to process and visualise SNP distances within a standalone single page application (SPA) built using React.js, AntDesign, and Cytoscope.js. Here, we describe and demonstrate the functionality of GraphSNP using examples from previously published datasets. GraphSNP is built as a simple, ready-to-use standalone visualisation tool to generate interactive charts, networks and trees to rapidly explore outbreak clusters and transmission using SNP distances and associated metadata. To fill this gap, we developed GraphSNP-A graph-based and interactive SNP distance visualisation tool for cluster and transmission analysis. In contrast, desktop and web-based visualisation tools such as Phyloviz and CATHAI, feature better interactivity but lack support for evaluating distance distribution and reconstructing transmission networks.

R packages, such as Outbreaker, Outbreaker2, and Regentrans, support both clustering and transmission mapping but require a high level of computer literacy and expertise in order to achieve readily interactive visualisations.
#React cytoscape software#
Ĭurrent tools that process and visualise SNP distance for outbreak analysis exist mainly as software libraries for programming language environments or lack the combined feature of clustering and transmission reconstruction. SNP distances generated from whole genome sequencing (WGS) provide the highest resolution for genotyping, and in combination with other epidemiological data allow investigators to decode transmission dynamics during outbreaks. Using Immutable allows for cheaper diffs, which is useful for updating graphs with many elements.The number of single nucleotide polymorphisms (SNPs) as the proxy of similarity between genomes-commonly referred to as SNP distance-are increasingly used as a measure of relatedness in outbreak studies. The props used to control the reading and diffing of the main props are listed below.Įxamples are given using Immutable. non JSON props), for example an object-oriented model or an Immutable model. This component allows for props of custom type to be used (i.e. The style attribute of the div containing CSS styles, e.g. The class attribute of the div containing space-separated class names, e.g. These props allow for setting built-in HTML attributes on the div created by the component that holds the visualisation: id If true, elements have immutable selection state, e.g. If true, nodes can not be moved at all, e.g. If true, nodes automatically can not be grabbed regardless of whether each node is marked as grabbable, e.g.
Whether shift+click-and-drag box selection is enabled, e.g. Whether the zoom level of the graph is mutable by user gestures (e.g. Whether the zoom level of the graph is mutable overall, e.g. The maximum zoom level of the graph, e.g. The minimum zoom level of the graph, e.g. Whether the panning position of the graph is mutable by user gestures such as swiping, e.g. Whether the panning position of the graph is mutable overall, e.g. Viewport mutability & gesture toggling panningEnabled


 0 kommentar(er)
0 kommentar(er)
